



Ploidy level of goldenberry, Physalis peruviana, plants obtained by anther culture
Nivel de ploidía de plantas de uchuva, Physalis peruviana, obtenidas mediante cultivo de anteras
Temas Agrarios
Universidad de Córdoba, Colombia
ISSN: 0122-7610
ISSN-e: 2389-9182
Periodicity: Semestral
vol. 27, no. 1, 2022
Received: 31 May 2022
Accepted: 02 June 2022

Abstract: The goldenberry, Physalis peruviana L., is an important fruit in Colombia due to its export value and nutritional quality. However, commercial crops face challenges of fruit heterogeneity and the presence of diseases that reduce fruit yield and quality. These drawbacks could be handled through different breeding methods to develop uniform cultivars, for example, through anther culture, which is used to rapidly produce homozygous lines. However, the ploidy level may change when using this technique. Therefore, in this study, the level of ploidy of parental and the plants obtained by anther culture was determined by cytogenetics, flow cytometry, and single-simple repeat or microsatellites (SSR). Additionally, the homozygous condition of obtained plants and the degree of heterozygosity of parental plants were evaluated using SSR. Cytogenetic analysis showed parental plants with 48 chromosomes and anther culture generated plants with 24, 32 and 48 chromosomes, and mixoploids and average nuclear DNA content between 5.04 and 20.08 pg. Diploid, tetraploid, hexaploidy, and octoploid plants were identified by flow cytometry; the highest levels of ploidy (6x and 8x) correspond to mixoploid plants found by cytogenetics. The SSRs did not allow identifying ploidy due to the lack of a particular band pattern, however, they showed heterozygosity in most of the plants obtained by anther culture. It was concluded that the anther culture modified the ploidy level with respect to the parental plants, and that flow cytometry is efficient, precise, and less laborious, compared to cytogenetics, to determine the ploidy level of goldenberry in the laboratory.
Keywords: Androgenesis, Cytogenetics, Flow cytometry, SSR.
Resumen: La uchuva, Physalis peruviana L., es una fruta importante en Colombia debido a su valor de exportación y calidad nutricional. Sin embargo, los cultivos comerciales enfrentan desafíos debido a la heterogeneidad de los frutos y a la presencia de enfermedades que reducen el rendimiento y la calidad. Estas desventajas podrían ser manejadas a través de diferentes métodos de mejoramiento para desarrollar cultivares uniformes, por ejemplo, mediante cultivo de anteras, la cual es empleada para producir líneas homocigotas. Sin embargo, el nivel de ploidía puede cambiar al usar esta técnica. Por lo tanto, en este estudio, el nivel de ploidía de las plantas parentales y las obtenidas por cultivo de anteras fue determinado por citogenética, citometría de flujo y secuencias cortas repetidas o microsatélites (SSR). Adicionalmente, la condición homocigota de plantas obtenidas y el grado de heterocigosidad de las plantas parentales fue evaluado usando SSR. El análisis citogenético mostró plantas parentales con 48 cromosomas y plantas generadas por cultivo de anteras con 24, 32 y 48 cromosomas, y mixoploides, y contenido nuclear promedio de ADN entre 5.04 y 20.08 pg. Mediante citometría de flujo se identificaron plantas diploides, tetraploides, hexaploides y octoploides, los niveles más altos de ploidía (6x y 8x) corresponden a plantas mixoploides encontradas por citogenética. Los SSRs no permitieron identificar el nivel de ploidía debido a la falta de un patrón de bandas particular y revelaron heterocigosidad en la mayoría de las plantas obtenidas por cultivo de anteras. Se concluyó que el cultivo de anteras modificó el nivel de ploidía con respecto a las plantas parentales, y que la citometría de flujo es eficiente,precisa y menos laboriosa, en comparación con citogenética, para determinar el nivel de ploidía de uchuva en laboratorio.
Palabras clave: Androgénesis, Citogenética, Citometría de flujo, SSR.
INTRODUCTION
Goldenberry, Physalis peruviana L., is the main export fruit in Colombia, with a national production in 2020 of 19,775 tons (t) and an average yield of 14.4 t/ha (Agronet, 2022). The export market is concentrated in the Netherlands, Germany, Belgium, Brazil, and Canada, which together purchase over 3,400 tons of fruit per year (ANALDEX, 2022). The commercial importance of goldenberry as an export fruit has resulted in a strong demand by producers for superior cultivars that are certified and carry genetic resistance to production-limiting factors, especially to the pathogen Fusarium oxysporum f. sp. physali (Foph), which causes vascular wilt and affects cultivated areas indiscriminately (Simbaqueba et al., 2018).
One breeding-acceleration strategy in a crop improvement program is the generation of homozygous lines by cultivation of isolated anthers or microspores. Anther culture allows the generation of haploid and doubled haploid (DH) plants, which can be converted directly into varieties or used as parental lines for the generation of hybrids (Jacquier et al., 2020). The generated haploid plants have the gametic number of chromosomes, while the doubled haploids carry the diploid chromosome number. Thus, the Colombian Agricultural Research Corporation undertook a project to produce homozygous goldenberry lines through anther culture (Suescun et al., 2011).
Goldenberry is cytogenetically variable, possibly because it is still an under-or semi-domesticated species. Menzel (1951) reviewed 25 species of the genus Physalis; they found 24 and 48 chromosomes and concluded that the basic number for the genus is x = 12. Therefore, plants with 48 chromosomes are tetraploid. However, variability for chromosome number has been reported by other authors; for instance, Bracamonte et al. (1997) reported diploid plants with 16 chromosomes for P. peruviana from Peru. Lagos (2006) counted 24, 32, 36,48, 52 and 54 chromosomes, besides mixoploidies. Rodríguez and Bueno (2006) found wild material with 24 chromosomes, and the commercial cultivars named ecotype Colombia and Kenya with 32 and 48 chromosomes, respectively. Bala and Gupta (2011) described polyploid cytotypes, with diploids, tetraploids and octaploids from India and other parts of the world and hexaploids found only in India. Franco-Florez et al. (2021) reported genotypes with chromosome number of 24, 48, 44 to 49 (aneuploids), and 36 to 86 (mixoploids).
Given the cytogenetic variability in goldenberry, the level of ploidy of donor or parental materials should be determined prior to initiating plant breeding via anther culture or through a hybridization program. Similarly, it is essential to know the level of ploidy of generated plants or offspring. The counting of chromosomes in somatic cells is the most direct and precise method to establish a karyotype and level of ploidy. This procedure allows for determination of chromosome number and structure (if chromosomes are sufficiently large), also for studying cell division. The basic steps in the treatment of mitotic and meiotic chromosomes include tissue collection, pretreatment, fixation, and staining (Bačovský et al., 2018).
Flow cytometry is an alternative technique for determining ploidy level. This process consists of the measurement of the fluorescence of stained nuclei, providing an estimate of the nuclear content of DNA within the somatic tissues of the plant (Adan et al., 2017). The interphase nuclear size is determined on a cytometer and a quantity of DNA is generated (C value).
The C value of experimental samples is compared with a known ploidy standard or target control. The ploidy of the DNA reported by cytometry is equivalent to the amount of DNA of a stable G1 phase chromosomal complement (Escobedo-Gracia-Medrano et al., 2018). Escobar et al. (2009) identified diploid, haploid and triploid anther derived plants by flow cytometry in Physalis ixocarpa. Larsen et al. (2018) used the GBS approach to estimate ploidy levels in apple and these results were consistent with flow cytometry. Likewise, Lattier et al. (2022) used chromosome counting, flow cytometry, and SSR to determine the ploidy of Corylopsis, a flowering shrub genus, and their results were consistent between methods, with clustering also based on ploidy.
In anther culture, in addition to determining the level of ploidy, it is necessary to know the degree of homozygosity. This can be done via molecular markers based on PCR such as microsatellites or simple-sequence repeats (SSR). These markers are one of the most informative for generation of molecular fingerprints, estimation of genetic diversity, molecular mapping, and marker-assisted improvement, due to their abundance, hypervariability and random distribution in the genome (Meena et al., 2020). In addition, they are codominant, meaning that the two alleles of a heterozygote can be identified (Cusaro et al., 2022). Thus, the aim of this work was to determine ploidy levels and zygotic condition of goldenberry plants derived from anther culture through three methods: chromosome counting, flow cytometry, and SSR, to select the most practical and accurate method to use in lab procedures.
MATERIALS AND METHODS
In this study, 15 donor or parental plants and 34 plants obtained from anther culture (Table 1) (Suescun et al., 2011) were analyzed by cytogenetics and SSR markers. The parental plants were collected in the towns of Arcabuco (05°45’3,06 N and 073°26’9,36” W; 2,643 masl) and Combita (05°41’47,5” N and 073°18’13,5” W; 2,830 masl) in the department of Boyaca, Colombia. Buds of parental plants were used as sources for in vitro plants via direct androgenesis.
Level of ploidy
The level of ploidy was determined by conventional cytogenetics and flow cytometry. Initially, the mitotic stage was established in the parental plants grown under greenhouse conditions at 23°C with 12 hours of light. For this, root samples were taken every hour for 24 hours and the number of cells in each phase of cell division was determined. Afterward, chromosome counting was performed in root tips using the methodology reported by Rodríguez and Bueno (2006) with some modifications. In the process, roots were pretreated with 0.25% colchicine for 2 hours at room temperature (~20°C). Then, roots were placed in deionized distilled water at 37°C for an hour. Next, acid hydrolysis was prepared, in 1N HCl for 25 minutes at room temperature. On a slide, root tips, approximately 2 mm in length, were stained with 2 or 3 drops of 2% propionic orcein for 15 minutes. Finally, the root was squashed using small strokes caused by the rubber of a pencil.
Flow cytometry analysis was requested from the University of Oregon, USA, for which in itro seedlings were sent. The internal process consisted in the isolation of nuclei from each plant and comparison with a diploid control, in this case, trout nuclei.
The DNA content of each genotype was estimated based on the relationship beween the average of the highest peak in the histogram and the mean peak G1 control, multiplied by five (DNA content of the control in picograms). The statistical relationship between the chromosome number and the amount of DNA was determined by regression analysis. The total variation among variables explained by the regression model was evaluated through the coefficient of determination, R2 (Steel et al., 1997).
Molecular characterization with microsatellite markers (SSR)
DNA was isolated from young leaves following the CTAB protocol (Rocha, 2002), with minor modifications, including DNA precipitation with 2-propanol rather than ethanol. The pellet was diluted in distilled water, cleaned with chloroform: isoamyl alcohol, and subsequently reprecipitated with sodium acetate and 2-propanol. DNA quality was verified visually on 1% agarose gels using a Horizon 58 horizontal electrophoresis chamber (Life Technologies) after staining with 0.1 μg/mL ethidium bromide. The concentration and purity of the DNA were determined by the UV 260/280 spectrophotometer (Beckman Du530).
The DNA was amplified with five specific P. peruviana microsatellite markers (SSR), labeled as 77, 123, 126, 138, and 146 (Simbaqueba et al., 2011). The amplification conditions used were: 1X PCR buffer; 2 mM MgCl2; 0.2 μM dNTPs; 0.2 μM primers; 0.05 U/ μl Taq polymerase, and 15 ng/μl DNA, for a final volume of 15 μL per reaction. The amplification cycle consisted of initial denaturation at 95°C for 3 minutes followed by 35 cycles of 95°C for 30 seconds, 54°C for 30 seconds and 72°C for 90 seconds, and a final extension of 72°C for 8 minutes. The PCR product was checked on 1% agarose gels stained with 0.1 μg/mL ethidium bromide and on 6% polyacrylamide denaturing gels, 5 M urea stained with silver nitrate (AgNO3). Finally, to evaluate the genetic relationships among genotypes, the allelic data were converted to binary data, 0 for absence and 1 for presence. The similarity matrix was calculated using the Dice coefficient (1945) using UPGMA (unweighted clustering method with arithmetic averages). In addition, a dendrogram was built with the NTSYSpc software (Numerical Taxonomy System for Personal Computer) version 2.02g (Rohlf, 2009).
RESULTS AND DISCUSSION
Cytogenetic analysis
In cytogenetic studies, the determination of the cell cycle allowed us to know the time in which the greatest number of cells were present in metaphase, known as mitotic time. At metaphase, the chromosomes are compacted facilitating the counting after inhibition of the mitotic spindle. In the present study, interphase comprised 95.1% of the cell cycle, which coincides with other publications in the same species (Liberato et al., 2014). The second stage, cell division, covered 4.9% of the cycle, and a constant activity was observed throughout the 24-hour cycle (Figure 1), a common occurrence in meristematic cells where there is active cell division. The period between 13 and 14 hours was the most appropriate to collect roots due to the greater number of metaphase cells found in this interval (Figure 1).
Mitotic activity has been observed in some species in the morning (Sangur et al., 2021; Putri et al., 2022). However, in goldenberry these results have been variable. For example, Rodriguez and Bueno (2006) reported 9 and 10 hours; Diaz and Gonzalez (2008) described the mitotic hour at 14 hours for seedlings maintained in photoperiod of 16 hours, while in seedlings maintained with continuous light the maximum peak was at 16 hours; Liberato et al. (2014) for in vitro plants found that the mitotic time occurs at 11 hours. Mitotic activity depends on light hours, temperature, and abiotic stresses (Mohammed et al., 2018; Qi and Zhang, 2020); consequently, the variation observed in the different studies could be due to these factors.
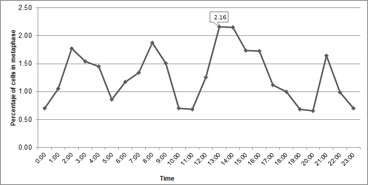
Therefore, to optimize the process of obtaining metaphase chromosomes, the conditions on which the mitotic time was determined in each study should be maintained.
Level of ploidy
The mitotic cell counts permitted visualization of 48 chromosomes (2n = 4x = 48) in the parental plants (Table 1). In the anther culture-derived plants there was variation in chromosome number; we found 24, 32, 36 and 48 chromosomes, and mixoploidies that ranged from 32 to 96 (Figure 2). The highest percentage of plants were those with 24 chromosomes (49%), followed by plants with 48 chromosomes (23%), mixoploids (16%), plants with 32 chromosomes (9%), and plants with 36 chromosomes (3%). According to Menzel (1951) the basic number of P. peruviana is 12, like other species of the family Solanaceae. For this reason, it is assumed that the parental plants are tetraploids. The plants having n = 2x = 24 would therefore be haploid, in which homozygosity would not necessarily be an obligatory condition produced by the anther culture technique, as seen in the Solanum tuberosum tetraploid potato (AboShama and Atwa, 2019). The homozygous condition, in this case obtained by anther culture, is a desirable state within breeding programs because it allows expression of positive or negative recessive characteristics, thus favoring the selection of new attributes. In goldenberry, according to the chromosome count, haploid and double haploid plants were obtained, which are the base to continue the breeding process through crosses for obtaining hybrids. Thus, the numerical compatibility of chromosomes is essential for choosing crossing parents and to guarantee the success of the offspring. Twenty-three percent of the obtained plants from anther culture were 2n = 4x = 48. Either spontaneous duplication, fusion of haploid nuclei, or production of plantlets from somatic tissue of the anther during embryogenesis may explain the presence of these diploid plants (Castillo et al., 2020). Nonetheless, observations made during the development of the embryos suggest that the all the plants obtained were derived from the microspores (Suescun, not published) since each embryo-like structure was quite independent from the anther wall. One way to clarify this phenomenon would be to compare the level of in vitro homozygous plants with the respective parental plants, in terms of segregation, under field conditions. Additionally, the use of informative SSR markers should allow for discrimination between homozygotes and heterozygotes.
| Generation* | Code | Chromosomenumber | Generation* | Code | Chromosome number |
| Parental | A3p | 48 | Parental | A10p | 48 |
| Generated | H36 | 34-36-38-42-56 | H17 | 24 | |
| H56 | 48 | H31 | 24 | ||
| Parental | A5p | 48 | H53 | 24 | |
| Generated | H21 | 24 | Parental | A13p | 48 |
| Parental | A7p | 48 | Generated | H6 | 24 |
| Generated | H3 | 48 | Parental | A16p | 48 |
| H10 | 32 | Generated | H41 | 48-52-56-64 | |
| H20 | 24 | Parental | A19p | 48 | |
| H24 | 24 | Generated | H45 | 48 | |
| H32 | 48 | Parental | B4p | 48 | |
| H33 | 56-70-80-82-96 | Generated | B4 | 48 | |
| Parental | A8p | 48 | Parental | B5p | 48 |
| Generated | H5 | 48 | Generated | B5 | 24 |
| H12 | 32 | Parental | EE1p | 48 | |
| H13 | 36-42-48-56-66-74-82 | Generated | G2 | 48 | |
| H14 | 36 | Parental | EE3p | 48 | |
| H19 | 48 | Generated | G1 | 32-44-46 | |
| Parental | A9p | 48 | G4 | 24 | |
| Generated | H42 | 24 | Parental | G-4p | 48 |
| Parental | A10p | 48 | Generated | K30 | 52-56-66-80 |
| Generated | H7 | 24 | K32 | 24 | |
| H8 | 24 | Parental | LE1p | 48 | |
| H9 | 24 | Generated | I1 | 48 | |
| H11 | 40-46-48-54 | I2 | 24 |
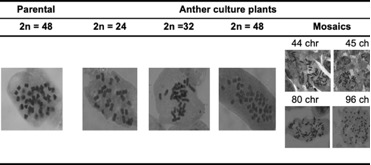
Sixteen percent of evaluated plants were mixoploids. This is an unusual condition, however, that could be caused by aberrant pairing, misalignment of the complete set of chromosomes, or chromosome duplication (Wang et al., 2018). Likewise, the observed mixoploidy could correspond to genetic instabilities associated with the regeneration of in vitro plants from undifferentiated calluses (Wang et al., 2018). In addition, other factors such as the type and concentration of growth regulators; the duration of the in vitro culture and the degree of endopolyploidy (duplication of chromosomes without subsequent karyokinesis and cytokinesis); as well as the type of explant employed might produce this condition (Pacey et al., 2019). Given that the plants used in this research were obtained from in vitro culture of anthers without the application of agents that induced chromosome duplication and without the induction of calluses, it is assumed that the different chromosomal contents could be related to the active process of cell division of the pollen grains in gametogenesis or to the stress generated in the anthers as a by product of pretreatment at different temperatures (Asadi et al., 2018; Lantos et al., 2022).
Flow cytometry analysis
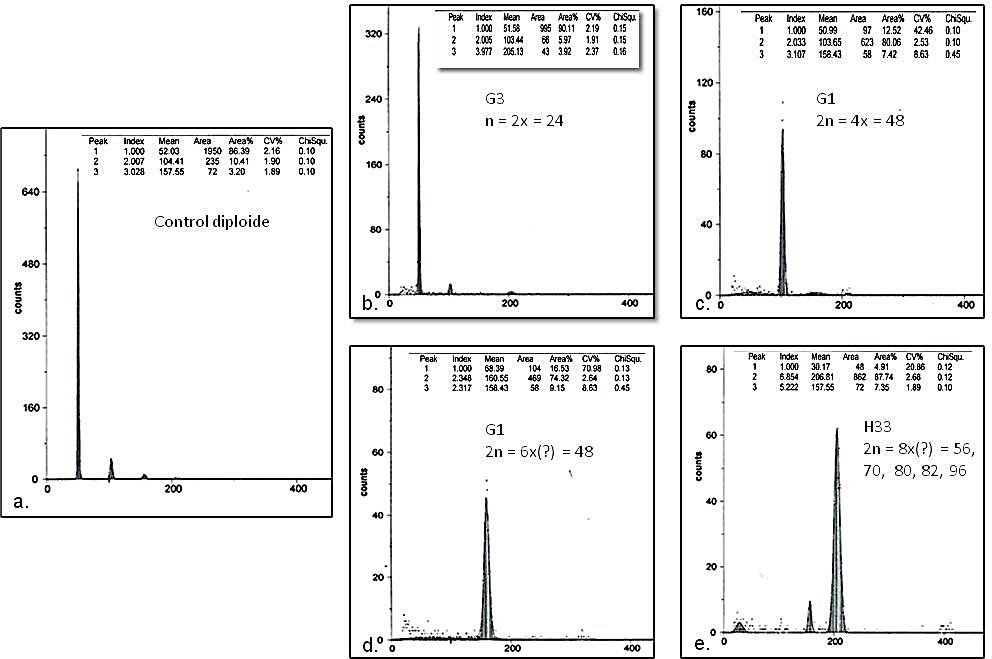
DNA content and ploidy level are important for both basic research and seed improvement and production (Dolezel and Bartos, 2005). In the histograms obtained by flow cytometry, diploid, tetraploid, hexaploidy, and octoploid seedlings were observed (Figure 3). The ratio between control and sample DNA was 1:1 in diploid plants; 1:2 in tetraploids; 1:3 in hexaploids and 1:4 in octoploids.
The average DNA content for the plantlets analyzed by flow cytometry ranged from 5.04 to 20.08 pg, with a coefficient of variation (CV) that varied between 1.92 and 5.64. The results by groups were as follows. In parental plants the average DNA content was 10.23 pg. The group of plants with 24 chromosomes presented the lowest variation (standard deviation) with an average DNA content of 5.13 pg; followed by plants with 48 chromosomes with an average of 11.99 pg; the group of plants with 32 chromosomes had
7.72 pg, while the group of mixoploids had the highest variability and an average of 15.06 pg (Table 2).
In this study, the average amount of DNA (2C) for goldenberry 2n = 4x = 48 was 10.23 pg (Table 2) which is significantly higher compared to model plants of the Solanaceae family. For example, in potato and tomato, 2n= 2x = 24, their DNA contents were 1.01 and 1.96 pg, respectively (Srisuwan et al., 2019; Zarabizadeh et al., 2022). These differences can be associated with amplification or reduction of repetitive DNA elements (Yan et al., 2021).
| Average parental | Average anther culture plants | 2n = 48 | 2n = 32 | n = 24 | Mixoploids | |
| DNA amount (pg) | 10.23 | 7.98 | 11.99 | 7.72 | 5.13 | 15.06 |
| Standard deviation | 0.13 | 4.52 | 3.23 | 3.63 | 0.07 | 7.10 |
In the set of 2n = 32 chromosomes the DNA content ranged from 5.15 and 10.29 pg; while in the mixoploid group it ranged from 10.03 and 20.08 pg. The genotype H33 was an atypical case, according to the cytogenetic study, since it has five predominant classes of chromosome numbers 56, 70, 80, 82, 96 (Table 1).
Despite this variation, in the histogram only one peak was observed (Figure 3e).
On the other hand, the ploidy level identified by flow cytometry and chromosome counting was consistent for plants with 24 and 48 chromosomes. We performed a regression analysis for plants with whole chromosome numbers, that is, excluding mixoploid plants,and a linear trend was observed between the amount of DNA and the number of chromosomes with an R2 of 0.72. The equation describing this relationship was y = 0.22x – 0.03, therefore, the amount of DNA increases by 0.22 as a chromosomeincreases. This allowed us to infer that there is a significant effect on the amount of DNA with respect to the number of chromosomes.
Although chromosome counting using roottip cells is the most accurate method for determining the level of ploidy in plants, this is arduous when a large amount of material is required to study. Therefore, the use of techniques such as flow cytometry contributes to decrease the complexity and labor, allowing to optimize the process of ploidy determination (Tomaszewska et al., 2021).
On the other hand, the ploidy level identified by flow cytometry and chromosome counting was consistent for plants with 24 and 48 chromosomes. We performed a regression
Molecular characterization with microsatellite markers (SSR)
Initially, SSRs 77, 123, 126, 138, and 146 were used to molecularly characterize the parental plants. SSRs 123, 126, and 138 were more informative to determine the homozygous or heterozygous condition since they amplified two or more bands, while SSR 146 amplified only one band and SSR 77 amplified four alleles of different sizes (220, 217, 215 and 210 bp). Most anther culture plants were heterozygous for the markers 123, 126, and 138. However, these plants have not yet been evaluated in the field to determine if androgenesis was from somatic cells or from pollen. If it were the case that plants come from somatic tissue, the plants should be identical to the parent plants.
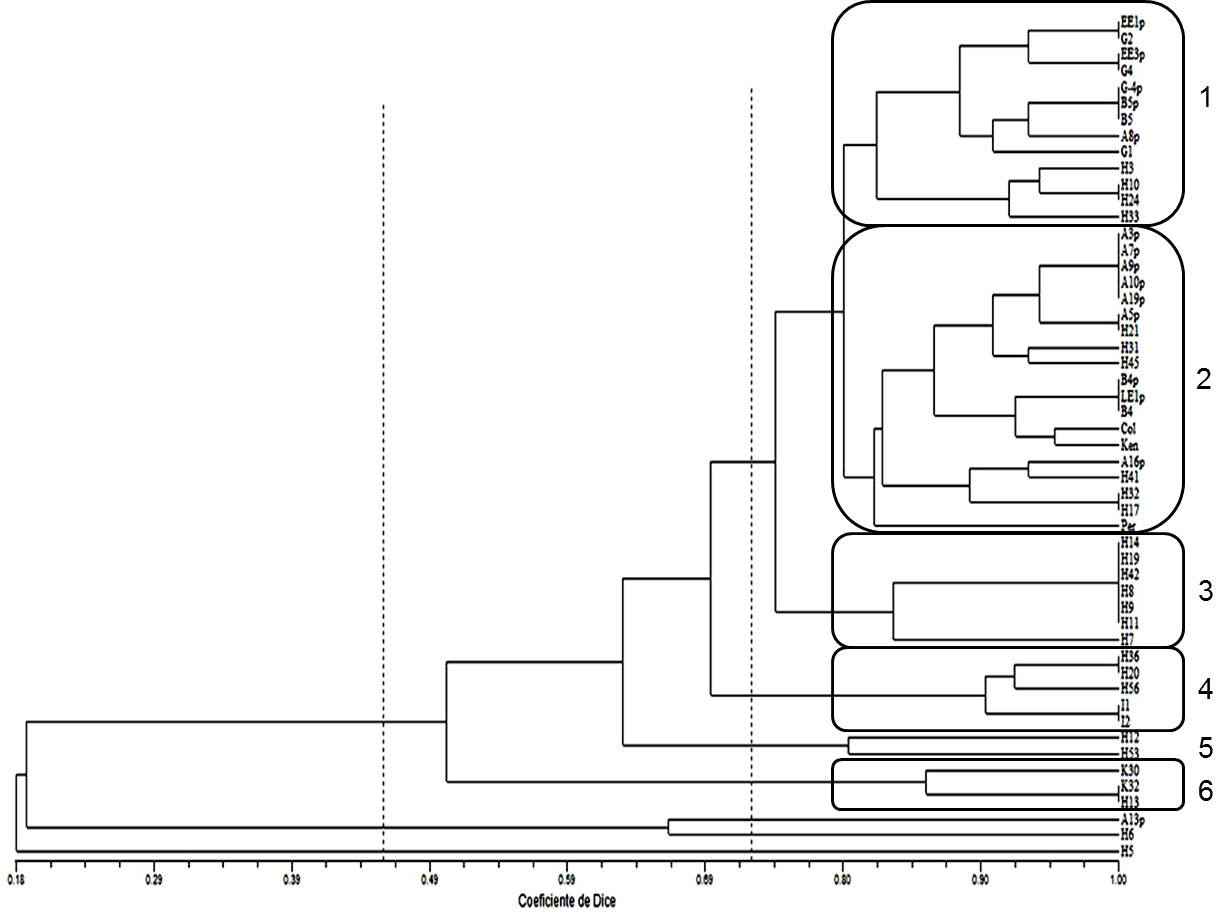
On the other hand, the SSRs were not useful to identify ploidy level since a specific band pattern was not observed according to the number of chromosomes. Likewise, the dendrogram, obtained from the UPGMA cluster analysis using the Dice’s similarity index (1945), did not show a clustering trend by ploidy level or by material origin (Figure 4). The parental plants were grouped mainly in groups 1 and 2, but next to them generated plants are also observed. An example of poor differentiation by ploidy level with these molecular markers is group 3 with seven plants, these are H14, H19, H42, H8, H9, H11 and H7. The chromosome number for them was 36, 48, 24, 24, 24, mixoploid and 24, respectively. Thus, in the dendrogram, we see that the first six plants were completely identical, which is the opposite of the number of chromosomes identified.
Most of the plants were found heterozygous by microsatellite analysis and might be originated by either diploid tissue of the anthers or by a phenomenon of non-reduced gametes or to the tetraploid condition of goldenberry (Asadi et al., 2018; Castillo et al., 2020). However, these plants may have developed from the sporophyte process of the microspores preserving the heterozygosity due to a possible tetrasomal inheritance. In addition, heterozygous plants with partial homozygosis would indicate genetic differences with the anther donor parent and therore would not necessarily originate from somatic cells (Asadi et al., 2018). Hypothetically, if tetrasomal segregation occurred, with four sets of chromosomes, each individual plant could contain between one and four different alleles. This was not observed for these markers, since the number of effective alleles was 1.6.
CONCLUSIONS
Generated plants from anther culture were diploid and double haploids with 24 and 48 chromosomes, respectively, and mixoploids with chromosomic numbers ranging from 32 to 96. The level of ploidy was estimated by flow cytometry and cytogenetics with consistent results between both techniques for plants with complete sets of chromosomes, but not for mixoploids. Finally, heterozygous generated plants were found, these plants were identified at the molecular level with microsatellites.
Acknowledgments
The authors thank MADR and Agrosavia for funding the research. In addition to Ligia Suescún for obtaining of plants from thee anther culture.
REFERENCES
AboShama, H.M. and Atwa, MM. 2019. Anther Culture in Potato (Solanum tuberosum L.) in vitro. J Plant Biochem Physiol. 7:244.
Adan, A., Alizada, G., Kiraz, Y., Baran, Y. and Nalbant, A. 2017. Flow cytometry: basic principles and applications. Critical Reviews in Biotechnology 37(2):163- 176.
Agronet – Ministerio de Agricultura de Colombia. 2022. Electronic search in May. https://www.agronet.gov.co/estadistica/ Paginas/home.aspx?cod=1
ANALDEX – Asociación Nacional de Comercio Exterior. 2022. Informe de las exportaciones de uchuva. Electronic search in May. https://www.analdex.org/2021/07/30/ informe-exportaciones-de-uchuva- mayo-2021/
Asadi, A., Zebarjadi, A., Reza Abdollahi, M and Seguí-Simarro, JM. 2018. Assessment of different anther culture approaches to produce doubled haploids in cucumber (Cucumis sativus L.). Euphytica 214: 216. https://doi.org/10.1007/s10681-018-2297-x
Bačovský, V., Hobza, R. and Vyskot, B. 2018. Technical Review: Cytogenetic Tools for Studying Mitotic Chromosomes. In: Bemer, M. and Baroux, C. (eds) Plant Chromatin Dynamics. Methods in Molecular Biology, vol 1675. Humana Press, New York, NY. https://doi.org/10.1007/978-1-4939-7318-7_30
Bala, S. and Gupta, RC. 2011. Effect of secondary associations on meiosis, pollen fertility and pollen size in cape gooseberry (Physalis peruvian L.). Chromosome Botany 6: 25-28. https://doi.org/10.3199/iscb.6.25
Bracamonte, O., Guevara, M., González, R., Cox, E., Siles, M. and Maguiña, E. 1997. Estudio citogenético de Physalis peruviana “capulí de la costa”. Revista Universidad Nacional de San Marcos.
Castillo, A.M., Valero-Rubira, I., Burrell, M.A., Allué, S., Costar, M.A. and Vallés, M.P. 2020. “Trichostatin A Affects Developmental Reprogramming of Bread Wheat Microspores towards an Embryogenic Route”. Plants 9(11): 1442. https://doi.org/10.3390/plants9111442
Cusaro, C.M., Grazioli, C., Zambuto, F., Capelli, E. and Brusoni, M. 2022. An Improved Method for Assessing Simple Sequence Repeat (SSR) Variation in Echinochloa crus-galli (L.) P. Beauv (Barnyardgrass). Diversity 14(3). https://doi.org/10.3390/d14010003
Diaz, D.E. y Gonzalez, D.C. 2008. Desarrollo de procesos organogénicos y estudio del efecto de la colchicina sobre yemas vegetativas de Physalis peruviana L. Tesis Biología, Universidad Pedagógica y Tecnológica de Colombia, Tunja
Dice, L.R. 1945. Measures of the amount of ecological association between species. Ecology 26 (3): 297-302. https://doi. org/10.2307/1932409
Escobedo-Gracia-Medrano, R.M., Burgos- Tan, M.J., Ku-Cauich, J.R. and Quiroz-Moreno, A. 2018. Using Flow Cytometry Analysis in Plant Tissue Culture Derived Plants. In: Loyola- Vargas, V., and Ochoa-Alejo, N. (eds) Plant Cell Culture Protocols. Methods in Molecular Biology 1815: 317-332. https://doi.org/10.1007/978-1-4939- 8594-4_22
Escobar-Guzman, R., Hernandez-Godinez, F., Martinez, O., and Ochoa-Alejo, N. (2009). In vitro embryo formation and plant regeneration from anther culture of different cultivars of Mexican husk tomato (Physalisxocarpa Brot.). Plant Cell, Tissue and Organ Culture. 96: 181–189
Franco-Florez, V., Liberato Guío, S.A., Sánchez-Betancourt, E., García- Arias, F.L. and Núñez Zarantes, VM. 2021. Cytogenetic and cytological analysis of Colombian cape gooseberry genetic material for breeding purposes. Caryologia 74(3): 21-30. https://doi.org/10.36253/caryologia-1081
Jacquier, N., Gilles, L., Pyott, D., Martinant, J-P., Rogowsky, P. and Widiez, T. 2020. Puzzling out plant reproduction by haploid induction for innovations in plant breeding. Nature Plants, Nature Publishing Group, 6(6): 610-619. https://doi.org/10.1038/s41477-020- 0664-9
Lagos, T.C. 2006. Comportamiento citogenético de Physalis peruviana. En: Biología reproductiva, citogenética, diversidad genética y heterosis en parentales de uvilla o uchuva Physalis peruviana L. Tesis de grado para optar al título de Doctor en Ciencias Agropecuarias, Universidad Nacional de Colombia, Palmira.
Lantos, C., Lehoczki-Krsjak, S. and Pauk, J. 2022. Induction of in vitro androgenesis in anther culture of recalcitrant einkorn (Triticum monococcum L.). Plant Cell Tiss Organ Cult 150: 417–426. https://doi.org/10.1007/s11240-022- 02293-6
Larsen, B., Gardner, K., Pedersen, C., Orgaard, M., Migicovksky, Z., Myles, S. and Toldam-Andersen, TB. 2018. Population structure, relatedness and ploidy levels in an apple gene bank revealed through genotyping-by- sequencing. PLoS ONE 13, e0201889 6093671 10.1371/journal.pone.0201889
Lattier, J.D., Ballard, H.E., Kramer, M. and Pooler, M. 2022. Genome size, ploidy levels, and development of novel SSR primer to evaluate genetic diversity of Corylopsis Siebold & Zucc. germplasm collections. Genet Resour Crop Evol 69, 2203–2216. https://doi.org/10.1007/s10722-022-01371-0
Liberato, S., Sanchez-Betancourt, Erika., Argüelles, J., González, C., Nunez, V. and Barrero. L. 2014. Cytogenetic of Physalis peruviana L., and Physalis floridana Rydb. Genotypes with differential response to Fusarium oxysporum. Revista Corpoica 15(1): 51-61.
Meena, R., Bhandari, M. and Ginwal, H. 2020. Usage of microsatellite markers for characterization of polyploids: A case study in reference to hexaploid bamboo species. Silvae Genetica 69: 94–97. https://doi.org/10.2478/sg-2020-0013
Menzel, M.Y. 1951. The Cytotaxonomy and Genetics of Physalis. Proccedings of the American Phylosophycal Society 95: 132-183
Mohammed, B., Farahi Bilooei, S., Dóczi, R., Grove, E., Railo, S., Palme, K., Anicet Ditengou, F., Bögre, L. and López-Juez, E. 2018. Converging Light, Energy and Hormonal Signaling Control Meristem Activity, Leaf Initiation, and Growth. Plant Physiology 176(2): 1365–1381.https://doi.org/10.1104/pp.17.01730
Pacey, E.K., Maherali, H. and Husband, B.C. 2019. The influence of experimentally induced polyploidy on the relationships between endopolyploidy and plant function in Arabidopsis thaliana. Ecology and evolution 10(1): 198–216. https://doi.org/10.1002/ece3.5886
Putri, ICS., Yuniastuti, E. and Parjanto, P. 2022. The rambutan (Nephelium lappaceum L.) chromosomes. Biodiversitas 23(4): 2196-2202. https://doi.org/10.13057/biodiv/d230455
Qi, F. and Zhang, F. 2020. Cell cycle regulation in the plant response to stress. Frontiers in Plant Science 10:1765. https://doi.org/10.3389/fpls.2019.01765
Redpath, LE., Aryal, R., Lynch, N., Spencer, JA., Hulse-Kemp, A.M., Ballington, JR., Green, J., Bassil, N., Hummer, K., Ranney, T. and Ashrafi, H. 2022. Nuclear DNA contents and ploidy levels of North American Vaccinium species and interspecific hybrids. Scientia Horticulturae 297: 110955. https://doi.org/10.1016/j.scienta.2022.110955
Rocha, P.J. 2002. Teoría y práctica para la extracción y purificación del ADN de palma de aceite. Palmas 23(3): 9-17
Rodríguez, N. and Bueno, M. 2006. Study of the cytogenetic diversity of Physalis peruviana L. (Solanaceae) L. Acta Biológica Colombiana 11(2): 75-85.
Rohlf, F.J. 2009. NTSYSpc Numerical taxonomy and multivariate analysis system. Applied Biostatistics Inc. New York. ISBN: 0-925031-31-3
Sangur, K., Smith, A. and Tomasoa, M. 2021. The Mitotic Index of Cajanus cajan from Kisar Island, in the Southwest of Maluku. Biosaintifika: Journal of Biology & Biology Education 13(2): 128-134. http://dx.doi.org/10.15294/biosaintifika. v13i2.29496
Simbaqueba, J., Catanzariti, A. M., González, C. and Jones, D.A. 2018. Evidence for horizontal gene transfer and separation of effector recognition from effector function revealed by analysis of effector genes shared between cape gooseberry- and tomato- infecting formae speciales of Fusarium oxysporum. Molecular Plant Pathology 19(10): 2302–2318. https://doi.org/10.1111/mpp.12700
Simbaqueba, J., Sanchez, P., Sanchez, E., Nuñez Zarantes, V.M., Chacon, M.I., Barrero, L.S. and Mariño-Ramírez, L. 2011. Development and Characterization of Microsatellite Markers for the Cape Gooseberry Physalis peruviana. PLoS ONE 6(10): e26719. ht t ps: / / doi. org/ 10. 1371/ journal. pone.0026719
Srisuwan, S., Sihachakr, D., Martín, J., Vallès, J., Ressayre, A., Brown, S.C. and Siljak-Yakovlev, S. 2019. Change in nuclear DNA content and pollen size with polyploidisation in the sweet potato (Ipomoea batatas, Convolvulaceae) complex. Plant biology (Stuttgart, Germany) 21(2): 237–247. https://doi.org/10.1111/plb.12945
Steel, R.G.D., Torrie, J.H. and Dickey, D.A. 1997. Principles and Procedures of Statistics. A biometrical approach. Third Edition, McGraw-Hill. New York. ISBN 0070610282
Suescun, L., Sanchez-Betancourt, E., Gomez, M., Garcia, F.L. and Nuñez, V.M. 2011. Producción de plantas genéticamente puras de uchuva. Corpoica, MADR, Novacampo, Cámara de Comercio de Bogotá. Editorial Kimpres Ltda. 44p.
Tomaszewska, P., Pellny, T.K., Hernández, L.M., Mitchell, R.A.C., Castiblanco, V., de Vega, J.J., Schwarzacher, T. and Heslop-Harrison, P. 2021. Flow Cytometry-Based Determination of Ploidy from Dried Leaf Specimens in Genomically Complex Collections of the Tropical Forage Grass Urochloa s. l. Genes 12(7): 957. https://doi.org/10.3390/genes12070957
Wang, GF., Qin, HY., Sun, D., Fan, ST., Yang, YM., Wang, ZX., Xu, PL., Zhao, Y., Liu, YX. and Ai, J. 2018. Haploid plant regeneration from hardy kiwifruit (Actinidia arguta Planch.) anther culture. Plant Cell Tiss Organ Cult 134: 15–28. https://doi.org/10.1007/s11240-018- 1396-7
Yan, L., Zhang, Y., Cai, G., Qing, Y.,Song, J., Wang, H., Tan, X., Liu, C., Yang, M., Fang, Z. and Lai, X. 2021. Genome assembly of primitive cultivated potato Solanum stenotomum provides insights into potato evolution. G3 Genes|Genomes|Genetics 11(10). https://doi.org/10.1093/g3journal/jkab262
Zarabizadeh, H., Karimzadeh, G., Monfared, S.R. and Esfahani, S.T. 2022. Karyomorphology, ploidy analysis, and flow cytometric genome size estimation of Medicago monantha populations. Turkish Journal ofBotany46(1). https://doi.org/10.3906/bot-2105-22
Notes

